RareVariantVis: a new tool for visualization of causative variants in rare monogenic disorders using whole genome sequencing data.
A bioinformatic tool made by Tomasz Stokowy at the UiB Genomic Core Facility.
Main content
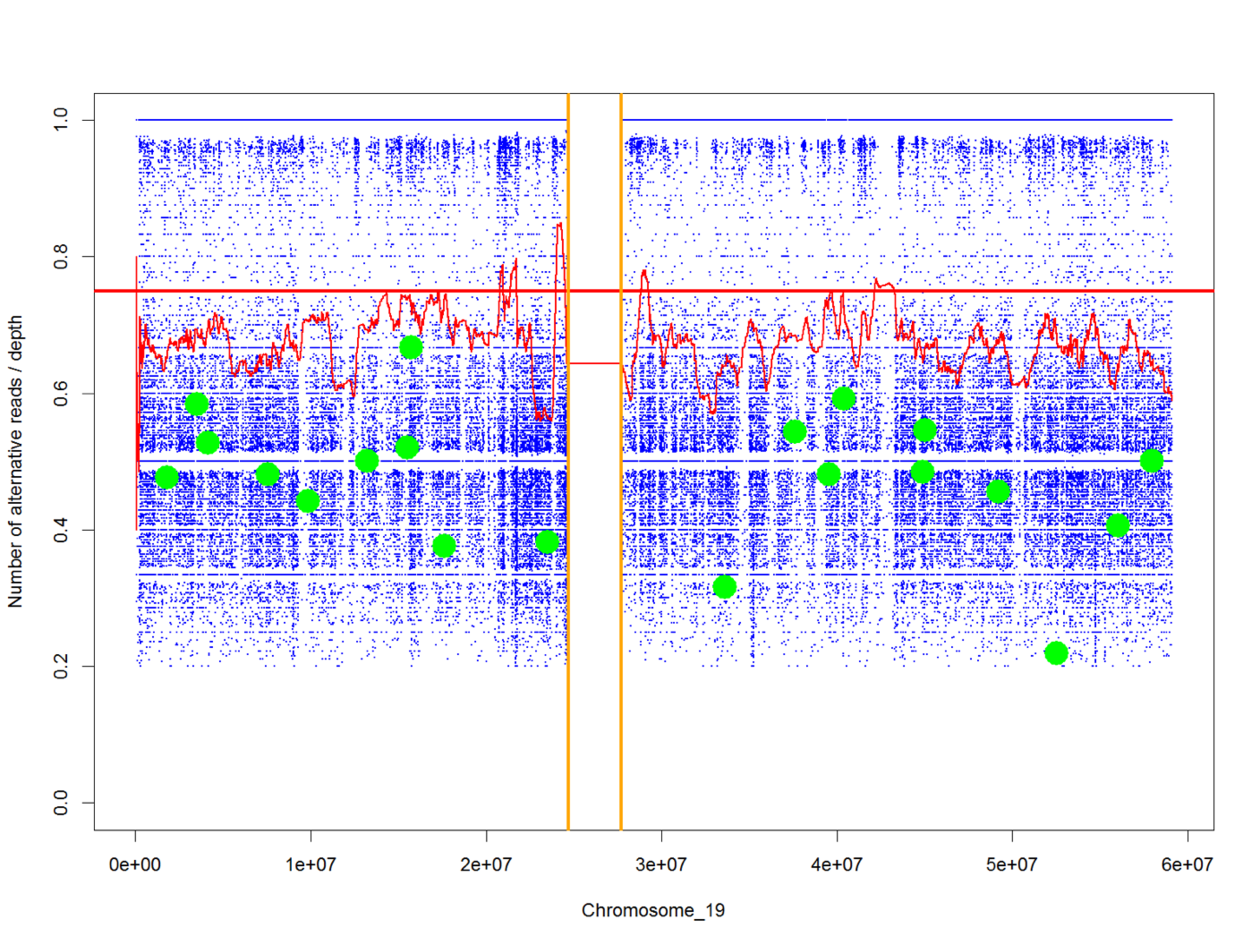
Genetic variants causing rare diseases can be identified by sequencing the genome or the exome (the part of the genome encoding genes) of the affected child together with the parents. Variants in the affected child not found in either of the parents (de-novo variants) or variants present in two copies in the child (and one in either of the parents) may cause disease. The RareVariantVis aids in the analysis of genome sequence data for both germ line and somatic variants. It provides a visualisation of the variants along the chromosomes and makes available all relevant information concerning positions, frequency, zygosity etc. De-novo variants and rare variants can be flagged in different colors facilitating identification of possible causative variants. The tool is available as a bioconductor package – and an updated version is being prepared.
RareVariantVis publication: Stokowy et al., Bioinformatics 2016