Protein function can be revealed by microscopy
This month, scientists from the NAT-group at the Department of Molecular biology (UiB) published their newest research in the journal Scientific Reports. They performed microscopy imaging on live yeast cells to study the function of human proteins.

Main content
Scientific Reports 6, Article number: 31627 (2016), doi:10.1038/srep31627
Even though most features differ tremendously, we do actually have a lot in common with Baker´s yeast. Does this mean that yeast can be applied as part of modern medical research to study humans?
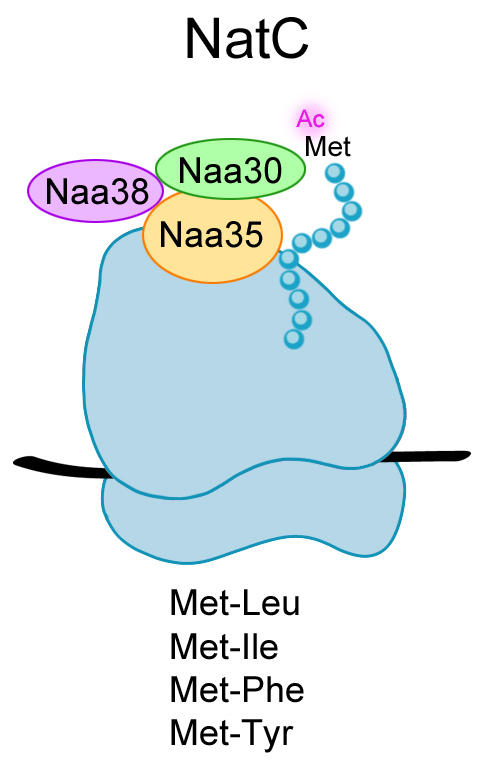
To date, six members of the NAT-family of enzymes have been identified, NatA-NatF, five of which also exist in yeast. Obviously, the enzymes have evolved differently but their most important property is conserved namely their ability to perform protein N-terminal acetylation. Even though more than 50 % of the cell´s proteins are modified in this way, scientists are still puzzled by the function of the acetyl group attached.
In this study, Camilla Osberg and colleagues investigated whether the human NatC complex (Figure 1) is able to N-terminally acetylate yeast proteins in vivo. Arl3, which is a known target of NatC, localizes to the Golgi apparatus when modified but mis-localizes to the cytosol in the absence of an acetyl group at its N-terminus (the beginning of the protein). This clear shift in subcellular localization of Arl3 from Golgi to cytosol represents active and inactive NatC, respectively. It was therefore applied as a localization model to study the function of NatC upon different combinations of yeast and human NatC subunits. In order to detect Arl3 in the cells by fluorescence microscopy, the protein was expressed with a GFP-tag fused to the C-terminus (the end of the protein). The catalytic subunit Naa30 of human NatC was found to restore the normal subcellular localization pattern of Arl3 in yeast cells (Figure 2). This microscopy-based localization model could be valuable to investigate whether mutation(s) in NatC subunits cause an abnormal function of the complex that entails disease.
The study was part of Camilla Osberg´s dissertation, which she defended earlier this year.
For more information on NAT enzymes and the group's work, please see our homepage.