Ruth Brenk
Professor, Associated member of the Computational Biology Unit
- E-mailRuth.Brenk@uib.no
- Phone+47 55 58 60 70
- Visitor AddressJonas Lies vei 915009 BergenRoom5A125B
- Postal AddressPostboks 78045020 Bergen
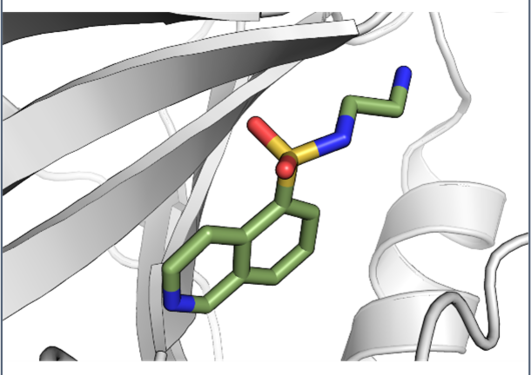
Our overall research goal is to improve methods used for structure-based drug design and to apply these methods to design inhibitors for enzymes with biological relevance. A key point in our research is the interplay of theoretical and experimental methods.
To read more about what we do, check out our homepage .
For a full list of publications click here.
Fields of competence
Research groups
Related content