MassShiftFinder
Blind search for post-translational modifications and amino acid substitutions using peptide mass fingerprints from two proteases.

Hovedinnhold
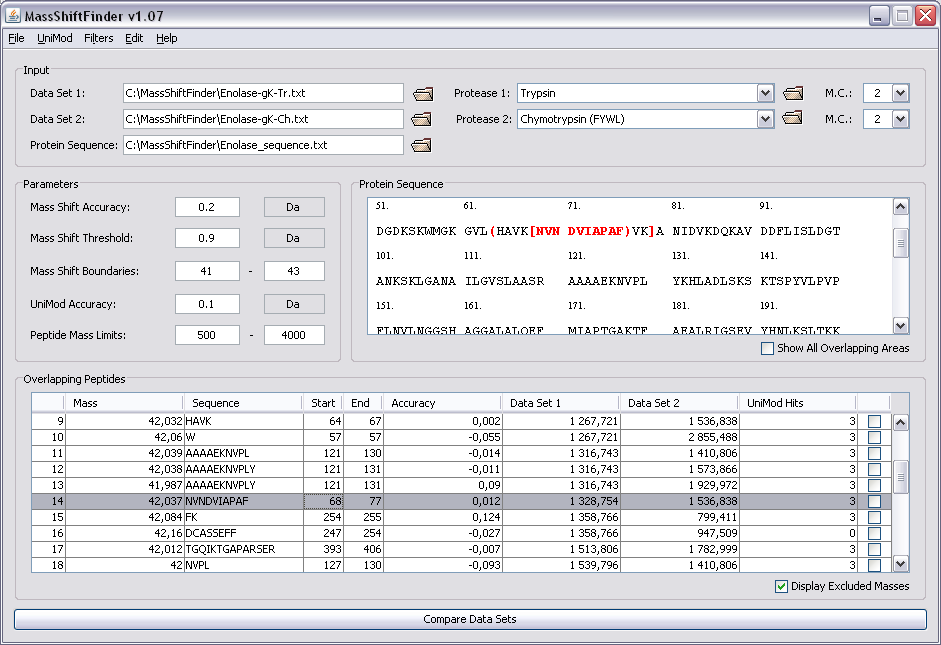
The identification of post-translational modifications and amino acid substitutions caused by mutations or nucleotide polymorphisms is an essential part of protein characterization. The most common way of doing characterization is by mass spectrometry, where the protein is digested by a protease creating a peptide mass fingerprint (PMF). There are two main approaches: (i) using a predefined set of possible modifications and substitutions or (ii) performing a blind search. The first option is straightforward, but detecting modifications or substitutions outside of the predefined set is not possible. Blind search does not have this constraint, and therefore has the potential of detecting both known and unknown modifications and substitutions. Combining the peptide mass fingerprints from two proteases result in overlapping sequence coverage of the protein, thereby offering alternative views of the protein and a novel way of indicating post-translational modifications and amino acid substitutions.
MassShiftFinder is a software tool for doing blind search using peptide mass fingerprints from two proteases with different cleavage specificities. The algorithm relies on overlapping peptides for the two proteases used, and can indicate both modifications and amino acid substitutions. The method can help restrict the area where the modification has occurred.
Download here: http://www.bioinfo.no/software/massShiftFinder