Paving the way for rational RNA-ligand design
Main content
Project summery
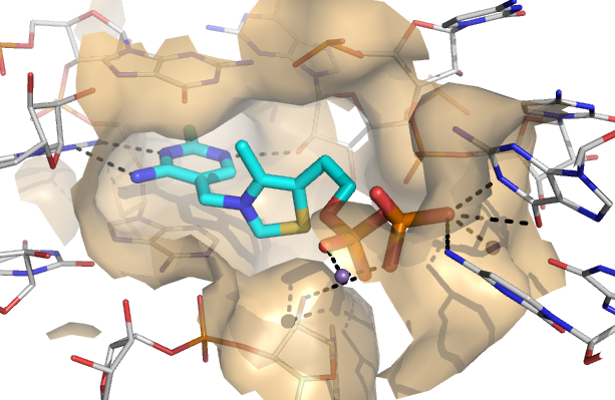
The vast majority of targets for approved drugs are proteins. In recent years, it has been increasingly realized that also RNA molecules constitute promising targets. However, compared to protein targets, RNA targets are vastly underexplored. By targeting RNA, the functions of currently undruggable protein-mediated pathways and the non-coding transcriptome can be modulated and thus the size of the druggable genome can be increased substantially. A major obstacle in RNA-targeting drug discovery is the lack of knowledge on how to obtain drug-like RNA ligands. The main goal of this project is therefore to establish a blueprint on how to design drug-like, potent, selective, and functional RNA ligands based on the 3D structure of the target and to advance our understanding of what drives potency and selectivity of RNA ligands on the molecular level. Focusing on RNA targets that contain druggable binding site will allow us to transfer methods that are at the forefront in structure-based design in the protein field to RNA. For that purpose, we have chosen two emerging targets for antibiotics, the FMN and TPP riboswitches (genetic switches in bacterial mRNA), as model systems. Hit compounds will be advanced through “design-synthesis-test” cycles and extensively characterized in terms of binding affinity and kinetics, functional activity, selectivity, and binding modes. This will deliver detailed knowledge about RNA-ligand interactions which is crucial to advance RNA-targeted drug discovery. In addition, the project will deliver advanced starting points for antibiotic drug discovery. We anticipate that this study will lead to a paradigm shift in the RNA field away from the currently rather unsuccessful “one size fits all” approach to a more target-centred approach where the nature of the binding site is taken into account. To achieve the ambitious goals, we have assembled an interdisciplinary team of experts in RNA biology, structure-based design, medicinal and organic chemistry, and drug discovery.
The team
In this project, a core group of four people will collaborate tightly:
- A researcher to carry out assay development, compound testing, and crystallography
- A postdoc to carry out compound synthesis
- A PhD student to carry out crystallography, design, and synthesis
- A PhD student to focus mainly on compound design
The researcher, the postdoc and one of the PhD students are already in place. Hiring of the second PhD student is ongoing. The project will be supervised by Ruth Brenk and Bengt Erik Haug.
Tasks of the PhD student to be hired
The main tasks of the PhD student to be hired will be to carry out structure-based design of FMN and TPP riboswitch ligands. One the one hand side, we have already discovered some ligands for these targets. The next steps will therefore be to design focused libraries around these ligands with the goal to improve their potencies. One the other hand, we also want to explore better the chemical space of FMN and TPP riboswitch ligands. To do so, the PhD student will carry out large docking screens to harvest the potential of chemically accessible virtual combinatorial libraries for RNA-ligand discovery. This will also entail to develop protocols on how to effectively navigate these large chemical spaces in a structure-based way. Depending on the interests of the student, the work can also include an experimental component.
This PhD project will be conducted in collaboration with the group of Prof. Jens Carlsson at Uppsala University in Sweden implying that this position will involve research stays in his lab.
Selected publications
- Rekand, I. H.; Brenk, R. DrugPred_RNA-A Tool for Structure-Based Druggability Predictions for RNA Binding Sites. J Chem Inf Model 2021.[Pubmed]|[DOI]
- Lundquist, K. P.; Panchal, V.; Gotfredsen, C. H.; Brenk, R.; Clausen, M. H. Fragment-Based Drug Discovery for RNA Targets. ChemMedChem 2021. [Pubmed]|[DOI]
- Daldrop P, Reyes FE, Robinson DA, Hammond CM, Lilley DM, Batey RT, Brenk R. Novel Ligands for a Purine Riboswitch discovered by RNA-Ligand Docking. Chem Biol 18 (3), 324-35 (2011). [Pubmed | DOI]
- Rekand IH, Brenk R. Ligand design for riboswitches, an emerging target class for novel antibiotics. Future Med Chem. 2017 Sep;9(14):1649-1662. [Pubmed | DOI | Post-print version]
- Wehler, T.; Brenk, R. Structure-Based Discovery of Small Molecules Binding to RNA. In RNA Therapeutics; Topics in Medicinal Chemistry; Springer, 2017; pp 47–77. [DOI]
- Daldrop P, Brenk R. Structure-Based Virtual Screening for the Identification of RNA-Binding Ligands. Methods Mol Biol 1103, 127-39 (2014). [Pubmed | DOI]